How Do Knowledge Graphs Contribute to Understanding COVID-19 Related Treatments?
Our Goals
 From 24 to 26 April 2020, the Scientific Data Management (SDM) group at TIB participated in the Pan-European hackathon #EUvsVirus organized with the aim of connecting experts, investors, and civilian organizations to devise together innovative solutions to the coronavirus outbreak. Citizens of all over the world responded to the call, and more than 2,160 proposals were submitted in 37 challenges. SDM joined efforts with the Software and Knowledge Engineering Laboratory (SKEL) at the National Center for Scientific Research “Demokritos” from Greece and presented Knowledge4COVID-19. SDM and SKEL aimed at showcasing the power of integrating scientific literature and biomedical databases to discover patterns that contribute to explaining the expected results of a corona-virus related treatment. Knowledge engineering techniques were followed to create a knowledge graph that comprises around 50,000 coronavirus-related scientific publications related through 52 million associations to more than 5 million biomedical entities like drugs, medical conditions, drug side effects, and interactions among drugs. Furthermore, machine learning methods allow for predicting potential adverse effects of drugs suggested for the treatment of COVID-19. Such estimations are quintessential to make safety decisions related to new clinical trials.
From 24 to 26 April 2020, the Scientific Data Management (SDM) group at TIB participated in the Pan-European hackathon #EUvsVirus organized with the aim of connecting experts, investors, and civilian organizations to devise together innovative solutions to the coronavirus outbreak. Citizens of all over the world responded to the call, and more than 2,160 proposals were submitted in 37 challenges. SDM joined efforts with the Software and Knowledge Engineering Laboratory (SKEL) at the National Center for Scientific Research “Demokritos” from Greece and presented Knowledge4COVID-19. SDM and SKEL aimed at showcasing the power of integrating scientific literature and biomedical databases to discover patterns that contribute to explaining the expected results of a corona-virus related treatment. Knowledge engineering techniques were followed to create a knowledge graph that comprises around 50,000 coronavirus-related scientific publications related through 52 million associations to more than 5 million biomedical entities like drugs, medical conditions, drug side effects, and interactions among drugs. Furthermore, machine learning methods allow for predicting potential adverse effects of drugs suggested for the treatment of COVID-19. Such estimations are quintessential to make safety decisions related to new clinical trials.
The Knowledge4COVID-19 Architecture
The SDM group developed a data-driven pipeline able to extract information about drugs and diseases from the scientific literature in the COVID-19 dataset, as well as relevant information for the extracted drugs (eg, drug-drug interactions, side effects, and indications ) from DrugBank. The extracted information is integrated into the Knowledge4COVID-19 knowledge graph. Figure 1 depicts the architecture of the Knowledge4COVID-19 data-driven pipeline.
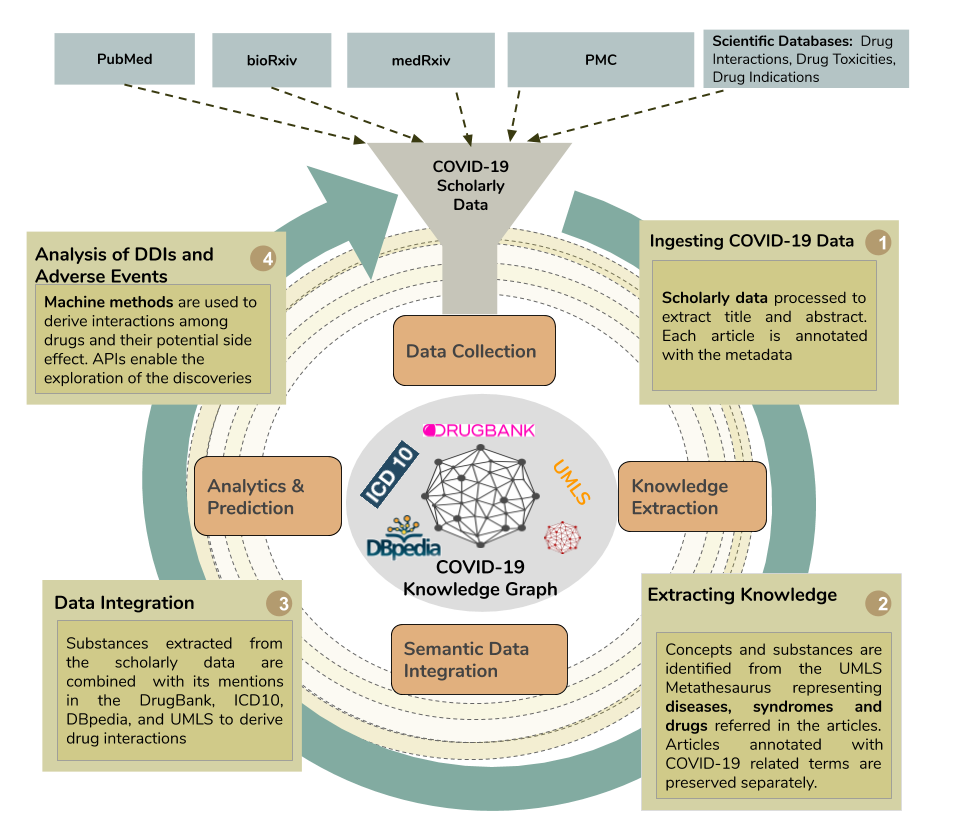
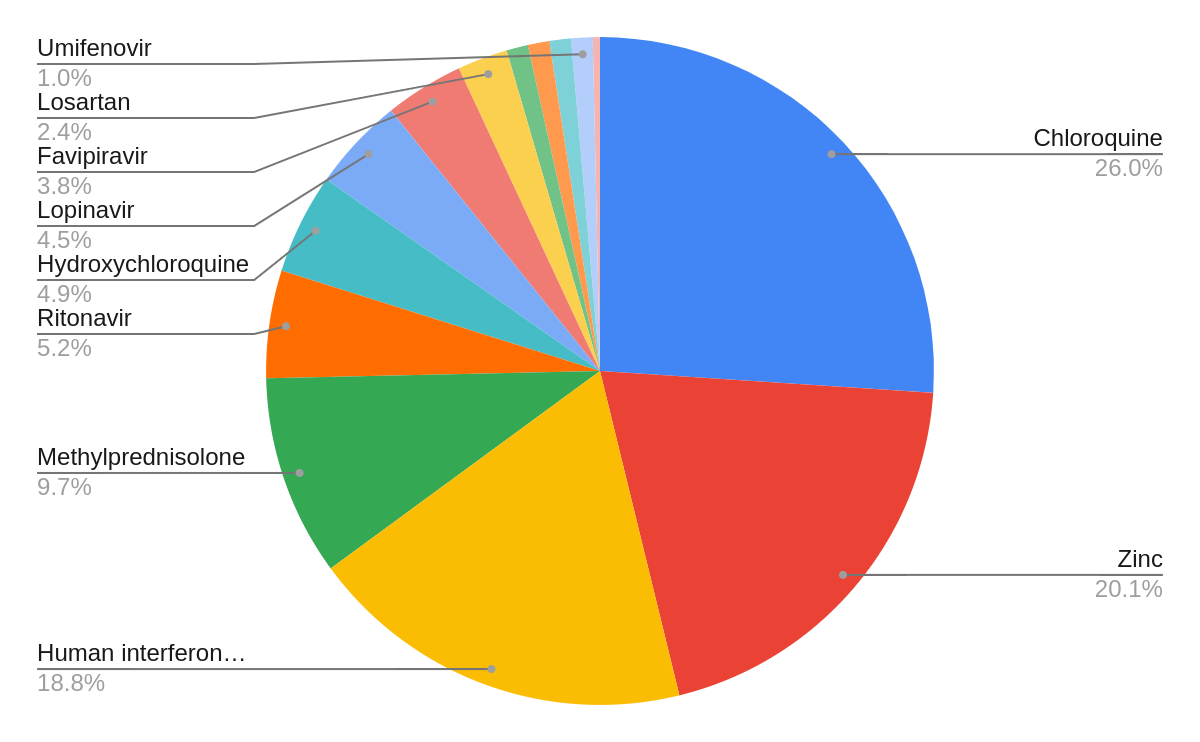
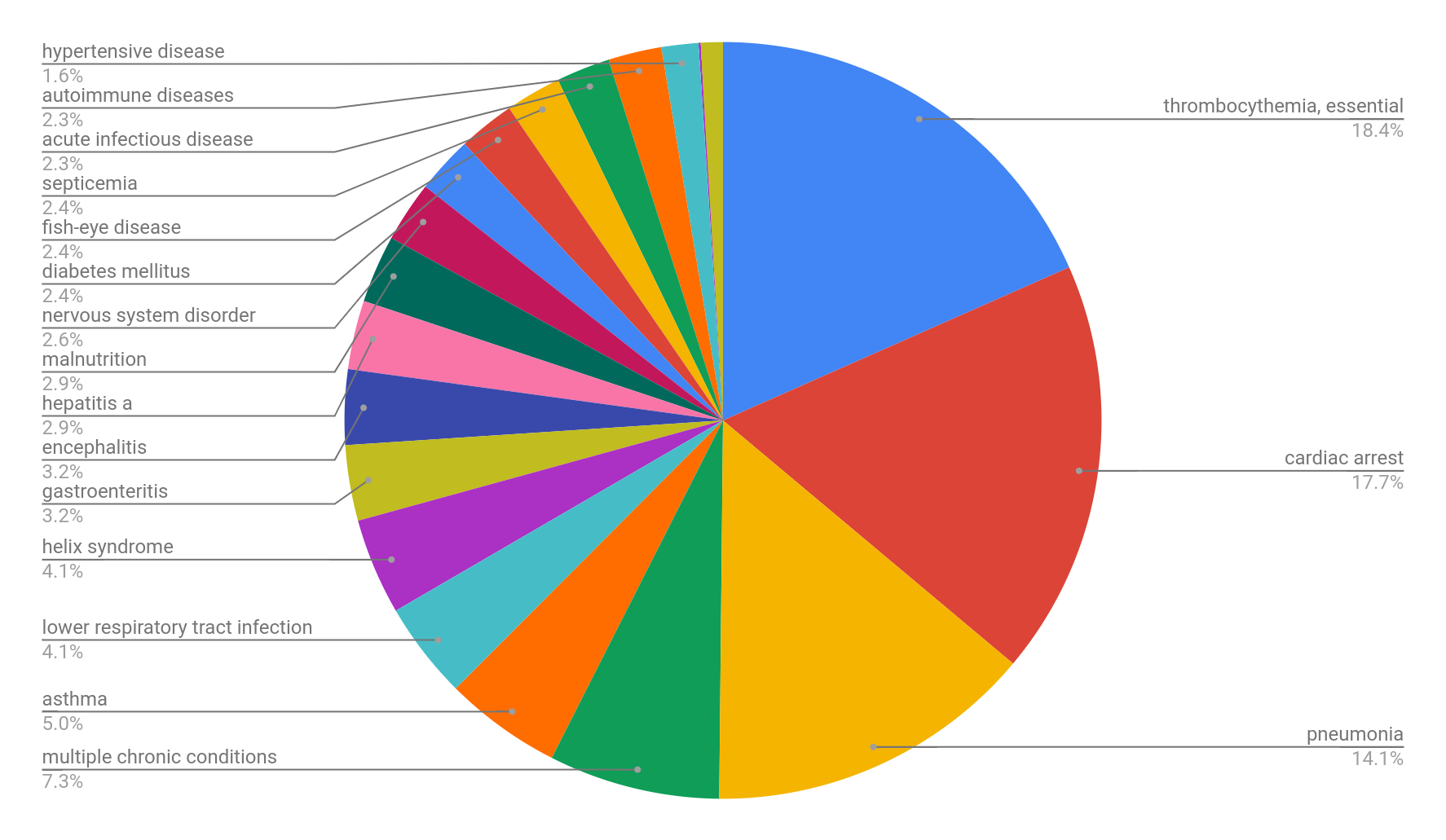
This dataset comprises 52,000 publications from Pubmed, bioRxiv, medRxiv, and PubMed Central (PMC). The natural language processing tool MetaMap was utilized to recognize drugs and diseases from the titles and abstracts of the integrated articles; the Unified Medical Language System (UMLS) was used to describe the extracted medical entities using a controlled vocabulary of medical terms. In total, 4,162 drugs and 2,012 medical conditions were extracted from the integrated publications. The coronavirus-related drugs are frequently mentioned in these publications; the drugs chloroquine, zinc, human interferon, methylprednisolone, ritonavir, hydroxychloroquine, and lopinavir represent the top most frequently mentioned drugs. Similarly, the most common medical conditions described in these publications include essential thrombocythemia, cardiac arrest, pneumonia, multiple chronic conditions, and asthma. Figures 2 and 3 report on the frequency distributions of COVID-19 drugs and related medical conditions. These distributions reveal interesting properties in the publications that composed the COVID-19 dataset.
The Knowledge4COVID-19 Knowledge Graph
The Knowledge4COVID-19 knowledge graph includes information about adverse effects that may exist when two or more drugs are taken together. Interactions between drugs that may cause adverse side effects have been extracted from the scientific database DrugBank. FALCON – an entity linking tool developed by the members of the SDM team – was used to extract this information from the textual descriptions in DrugBank. As a result, 2,205,099 drug-drug interactions and 5,965 drug toxicities are part of the Knowledge4COVID-19 knowledge graph. Furthermore, the SKEL team contributed with machine learning methods that resort to contextual information from the scientific literature stored in the Knowledge4COVID-19 knowledge graph to predict drug-drug interactions. In total, 22,346 potential novel drug-drug interactions are part of the Knowledge4COVID-19 knowledge graph. Knowledge4COVID-19 is linked to existing knowledge graphs to include encyclopedic and factual knowledge about drugs and conditions represented in DBpedia, Bio2RDF, and DrugBank (14,524). In-house tools developed by the SDM team (eg, SDM-RDFfizer) were used for knowledge graph creation. Knowledge4COVID-19 is publicly available via a SPARQL endpoint.
Patterns Among Drugs and Drug-Drug Interactions
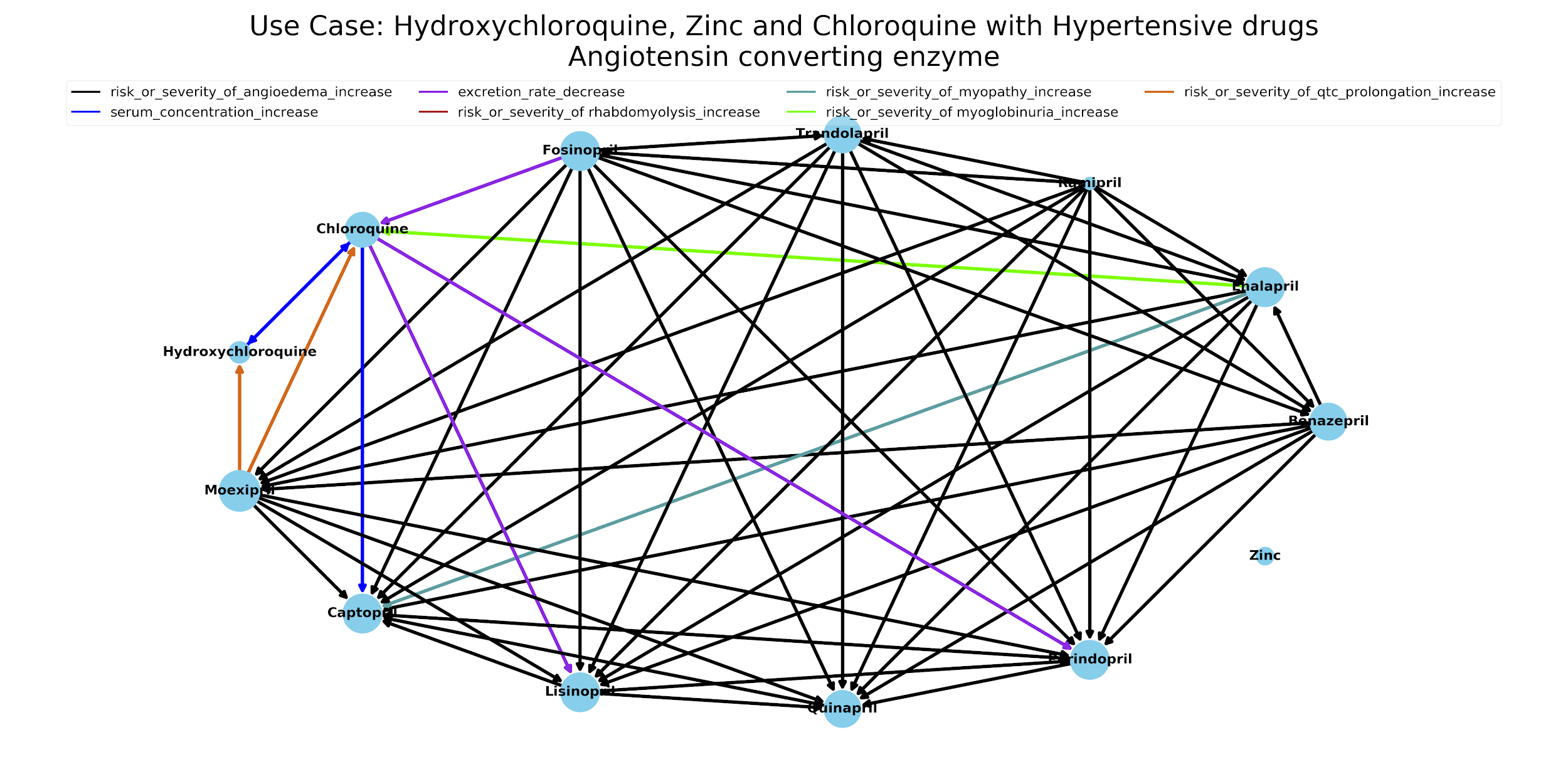
The representation of information in Knowledge4COVID-19 allows for uncovering patterns that facilitate the explanation of a treatment results. Furthermore, these interactions – in combination with the information about coronavirus-related drugs and medical conditions extracted from scientific articles – enable the understanding of the potential adverse medical conditions that may occur in the presence of conditions such as high blood pressure, asthma or diabetes. Figure 4 depicts the large number of drug-drug interactions that may exist – according to DrugBank – among coronavirus-related drugs and hypertensive drugs. Each edge in the graph that goes from a drug A to a drug B, represents that A affects B in some way; the colors of the edges represent the types of the drug-drug interactions extracted from DrugBank. As observed in this Figure, there is a large number of potential drug-drug interactions among this group of drugs. The analysis of these patterns embodies valuable information to assemble clinical trials and prescribe conscientious treatments. Knowledge4COVID-19 provides Web services that allow for visualizing all the drug-drug interactions among any group of drugs.
Next steps
In the future, we plan to integrate clinical COVID-19 related data to detect patterns that can explain correlation between survival, and drug interactions and toxicities. We also plan to connect the Knowledge4COVID-19 knowledge graph with the Open Research Knowledge Graph (ORKG). In addition, other types of interactions, eg, drug-target, protein-protein, and drug-side effects will be extracted from the literature and from scientific databases.
Our team
The Knowledge4COVID-19 team is composed of the following members:
- SDM: Anery Patel, Ariam Rivas, Ahmad Sakor, Vitalis Wiens, and Maria-Esther Vidal (Technical team), and Gabriela Ydler (dissemination).
- SKEL: Kostantinos Bougiatiotis, Fotis Aisopos, Anastasia Krithara, and George Paliouras


0 Antworten auf “How Do Knowledge Graphs Contribute to Understanding COVID-19 Related Treatments?”