Organizing COVID-19 research with the Open Research Knowledge Graph
by Allard Oelen and Markus Stocker
The COVID-19 crisis is driving substantial research with new articles published daily. To support COVID-19 research, many publishers decided to open access related articles. While access is crucial, organizing information published in articles is essential for effective research, but is extremely time consuming, and time is an asset that under these circumstances is as valuable as ever.
The Open Research Knowledge Graph (ORKG) aims to increase the efficiency of scholarly knowledge organization by representing knowledge in a structured manner thus making knowledge published in the scholarly literature more machine actionable and ultimately FAIR.
We present how the ORKG can organize the COVID-19 basic reproduction number, also called R0 of a COVID-19 infection. This number refers to the expected number of people that an infected person will infect. Thus, it indicates how contagious COVID-19 is. Together with the case fatality rate, R0 provides insights on how dangerous an infectious disease is. These numbers can differ across locations, because of population density or cultural differences. R0 numbers of some well-known diseases include 12-18 for measles, 2-5 for SARS, and 2-3 for the Common Cold.
Many articles have already been published that report COVID-19 R0 estimates. Being scattered across growing literature, it becomes increasingly expensive to organize R0 information, their value, confidence intervals, study location and time frame.
Traditionally, review articles organize the literature for a particular problem. As such, review articles are often very valuable but they have at least two shortcomings. First, just as original research articles, their content is unstructured and is thus hardly machine actionable. Second, they reflect the state of knowledge at a particular point in time.
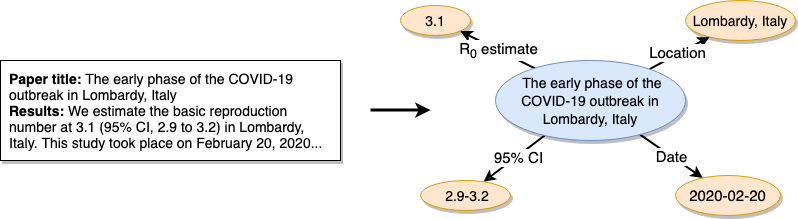
This is where the ORKG comes in. ORKG supports the structured description of scholarly work. For R0 research, the following information is particularly relevant: R0 estimate, a 95% confidence interval, the time frame and location of the study and (optionally) the methods used to determine the R0. In articles, this information is published as natural text, making the (human or machine) extraction of precisely this information rather difficult. With the ORKG we can publish the same information in a structured manner. As depicted by the following image, we thus translate information in natural text into corresponding machine actionable representations.
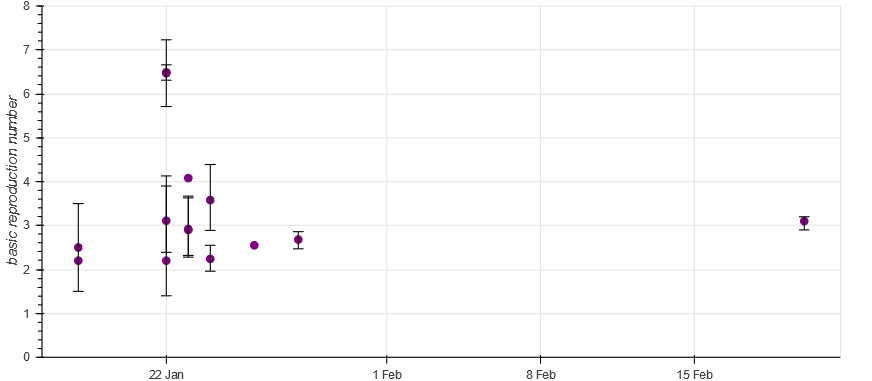
Machine actionable scholarly knowledge opens a range of very interesting possibilities. For instance, it is possible to automatically create literature overviews (or literature comparisons). The following picture showcases this for our use case on the COVID-19 basic reproduction number. Check it out.
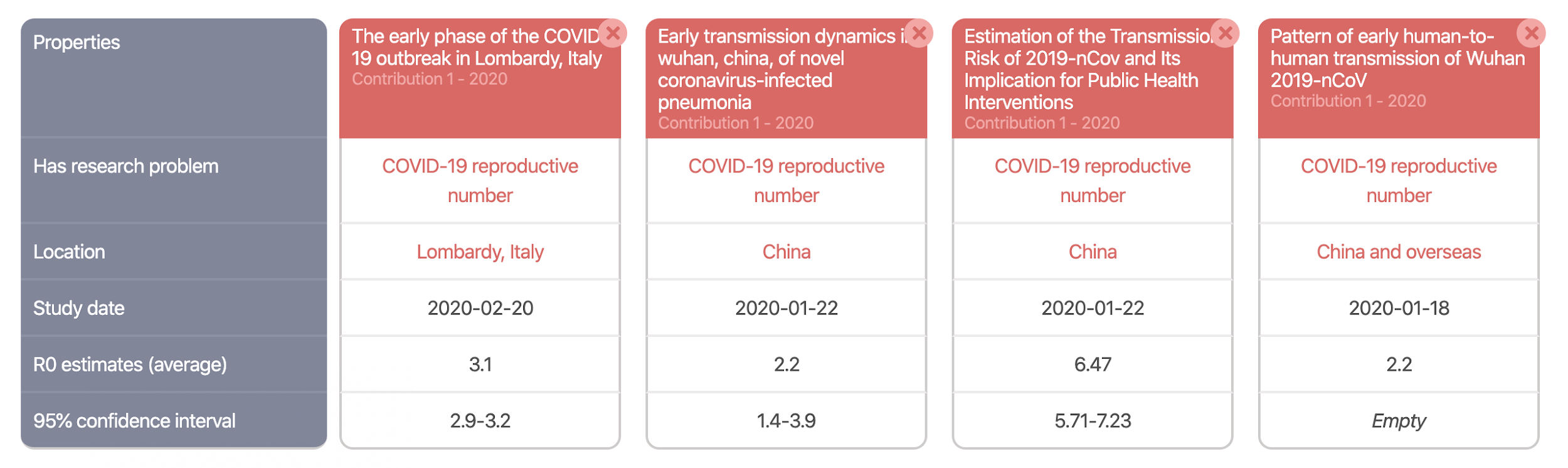
It is the structured representation of scholarly knowledge that enables the automatic creation of such overviews in ORKG. This is great, but only one interesting possibility. The other is that, contrary to review articles, ORKG overviews can evolve. As new literature on R0 research is published, it is straightforward to extend such an overview, which therefore continues to reflect in a comparable manner the current state of knowledge.
The real power of such ORKG overviews, however, can be seen if they are taken as data sources. Indeed, thanks to machine actionability of both the data and the data exchange protocol (REST API), it is possible to link the ORKG and overviews, specifically, with downstream data science. We demonstrate this by connecting Jupyter with ORKG to show how we can leverage the flexibility of data science environments and programming languages such as Python and R to visualize or otherwise process the COVID-19 comparison data. The following figure shows a possible result. Check it out.